Overview of TMAJ
What is TMAJ?
TMAJ is open-source software
that provides ways to store and retrieve information
related to tissue micro-arrays.
TMAJ was created by the Johns
Hopkins TMA Core Facility. The J in TMAJ refers to
Java,
as TMAJ is programmed 100% in Java.
Data Stored in TMAJ
TMAJ allows
the storage of a
wide variety of information related to TMA samples, including:
- Images on the ArraySlide
- Patients
- Specimens
- Donor Blocks
- Array Cores
- Array Blocks
- Array Slides
- Machine-Scored Data
- Image Analysis Data
Dynamic Database Structure
One of TMAJ's most powerful features is its
dynamic database
structure. This allows users with
administrative access to add custom fields for different organ systems. For example, a group studying prostate cancer
will record different data than another group who studies colon cancer. With this structure, an administrator can
have multiple organs, with each organ having its fields.
For example, a prostate type may have a field
called "Gleason Score". TMAJ
provides an application that allows an administrator to add, change, or
delete
these fields. Administrators may also
assign what choices a field may have.
For example, an administrator may assign the field "Primary
Gleason
Score" choices "1", "2", "3", "4",
and "5".
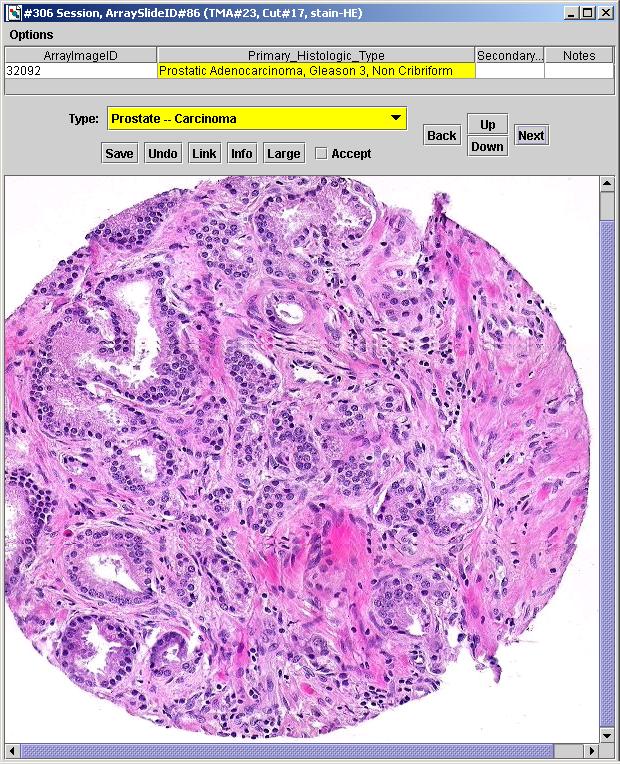
|
|
A
screenshot from the Images Manager application in
TMAJ.
In the above image, the user chooses the type
"Prostate -- Carcinoma". Based upon this choice, certain fields
and choices are displayed in the table. In the above image, the
field
displayed is "Primary_Histologic_Type". One
of its choices is "Prostatic
Adenocarcinoma, Gleason 3, Non Cribriform". This field and choice
are
only displayed when the user chooses the "Prostate -- Carcinoma"
type. If the user would have chosen the "Bladder" type, this
choice would not have been displayed.
|
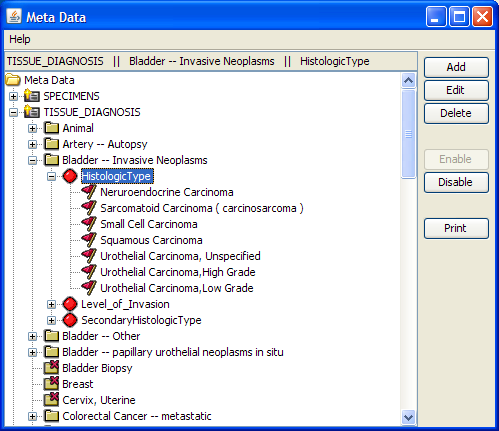
|
|
A
screenshot from the Meta
Data application in TMAJ. The Meta Data Application allows an administrator to
determine which types (e.g. prostate, bladder, etc) TMAJ will contain,
and what
fields each type will have. For example,
the "prostate" type may have a "Prostate_Zone" field, whereas
this would be irrelevant for a "bladder" type. Additionally,
each field may have choices
associated with it. In the above
example, we see the "Prostate Zone" field has choices such as
"Central Zone" and "Peripheral Zone".
|
Data Security & HIPPA
TMAJ has extensive security features, as it was designed with HIPPA
legislation in mind. These security features are default with a
new TMAJ installation, and can be fine-tuned by an administrator.
All users in TMAJ are assigned a username and password and are required
to log on to perform various functions. One of the
main types of protected data is patient information. This
includes protected patient data such as birth dates and names, as well
as limited-data-set identifiers such as surgical pathology
numbers. A TMAJ administrator may decide to deny all all access
to patient information outright, or may allow access to a subset of
specimen records. Access to other data types can also be tightly
controlled by an administrator. These other data types include
blocks, array-blocks, array-cores, array-images, array-slides,
image-analysis data, and meta-data. Different levels of
data-access may be provided, such as providing write access and
read-only access.
TMAJ also supports a concept of group-administrators, who have
adminative rights over a subset of users. Group-administrators
control only a subset of TMAJ data, and may assign privileges to this
data to the subset of users they control.
Image Analysis
TMAJ has capabilities for performing automated image analysis.
This was done by integrating the Frida image analysis package.
Applications of TMAJ
TMAJ has 5 major applications:
- SpecimensManager -- Used
to
enter data about Patients, Specimens and Donor-Blocks
- ArrayBuilder --
Used to design ArrayBlocks
- ArrayManager -- Used to
enter information about ArrayBlocks, ArraySlides, & Scans
- Import -- Used to
import ArrayImages from a Scanning Machine (like the BLISS or ACIS).
- ImageManager-- Used to
score
Images on an ArraySlide and perform image analysis.
- Meta Data -- Used to add
custom data fields to TMAJ.
- Administrator -- Used to
set security options
Overview of Technology Behind TMAJ
The client uses the server in 2 different ways:
- To read or write information to the database
- To get an image file from a Tissue MicroArray
Clients can read or write information to the database by first
contacting the Apache Tomcat server. TMAJ has a Java
servlet running on Tomcat. The client sends a Request class to
the Tomcat servlet and receives a Response class back. In the
Request class, the client places a username and password. Tomcat then
returns a Response back to the client based upon the permissions of the
person's user account. This Response class contains
information from the database. By requiring each Request class to
have a username and password, we can ensure that users only see
information for which they have been assigned access.
If the client sent a Request class which asked Tomcat to write
information to the database, the Response returned may only indicate if
the data was written successfully or failed. Users are prevented
from overwriting data to which they do not have access.
The Java servlet connects to the database using JDBC (Java Database
Connectivity). The TMAJ database may be your choice of MySQL,
Microsoft SQL Server, Oracle, or Sybase.
Image files are created by a technician using a scanning machine.
TMAJ supports the ARIOL from Applied Imaging, the BLISS from Bacus
Labs, the ACIS from Clarient (formally called ChromaVision), and the
ScanScope from Aperio Technologies.
A technician scans an array-slide, and the scanning machine converts
each spot on the array-slide into a jpeg image file.
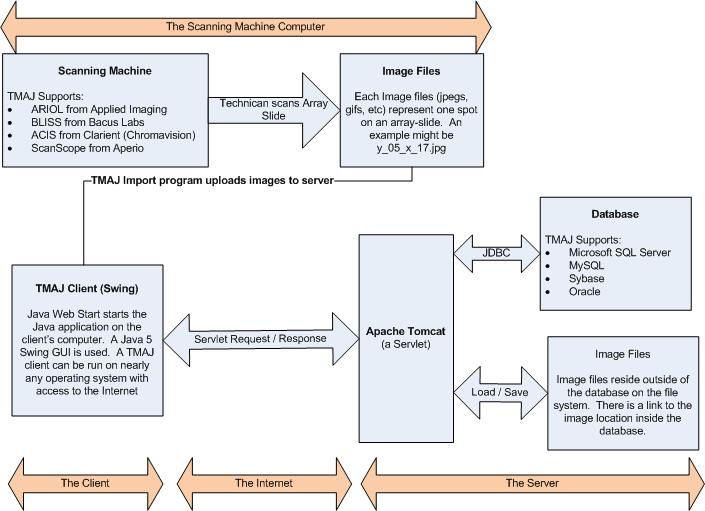
|
|
How the TMAJ
client and server interact, and the
technologies behind them.
|
Authors
TMAJ was created by Dr.
Angelo De Marzo, Dennis Faith, and Helen Fedor
of Johns Hopkins University, and
James Morgan, Brian Razzaque, and Joe Zimmerman of Vision Multimedia Technologies (VMT).
License
TMAJ is open source software; you can redistribute it and/or
modify it under
the terms of the GNU General
Public License as published by the Free Software Foundation, either
version 3 of the License, or (at your option) any later
version. This program is distributed in the hope that it
will be useful, but WITHOUT ANY WARRANTY; without even the implied
warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR
PURPOSE. See the GNU General
Public License for more details.
<< Back to the Manual
© Copyright 2009 |
All Rights Reserved | The Johns Hopkins University

